Evolutionary theory is often described in heroic terms as one of the great intellectual accomplishments of all time. Yet it is also often described in more humble terms as a “toolkit” for understanding how nature works. Most evolutionists do not go to work every morning with the final movement of Beethoven’s 9th playing in their heads. Instead, they show up at their jobs like a carpenter or plumber, size up the situation, and reach for the appropriate tools from their toolkit to get the job done.Some of these tools are physical—petri dishes for microbiologists, butterfly nets for entomologists, and the reagents and instruments of molecular biologists. But the most important tools in the evolutionary toolkit are conceptual—certain ways of thinking that can be used to study any property of any living creature. It is the conceptual toolkit that gives evolutionary theory its amazing generality.If you want to learn a craft, then observe a master craftsman. That label certainly fits Richard Lenski, who has become world famous for presiding over the longest running evolutionary experiment of all time, on the bacterium E. coli, which has now exceeded 65,000 generations. Lenski’s research has been widely reported in the popular media. My reason for interviewing him for This View of Life is not only to showcase his research, but also to show how his research illustrates the concept of evolutionary theory as a conceptual toolkit.DSW: Greetings, Rich, and welcome to This View of Life. You are much in demand and thanks for agreeing to this interview.RL: My pleasure, David. I enjoy these wide-ranging and open-ended opportunities to talk about science and evolution.DSW: I’d like to begin with some personal details. Your father was a sociologist (Gerhard Lenski) and your mother was a poet (Jean Lenski). How did you become an evolutionist?RL: Like a lot of choices we make, it didn’t happen all at once. Both of my parents grew up in religious families, and we went to church every week while I was growing up. However, my parents not only accepted the reality of evolution, they were fascinated by it. It influenced both my father’s work on human societies and my mother’s poetry. So I was exposed to evolutionary thinking as a kid, and in a positive way. On the other hand, I was not a naturalist. I loved playing sports and hiking, but I was not one of those kids into snakes or birds or bugs.I didn’t particularly like my high-school biology class. But when I was in college, I took a non-majors course that was team taught by the small biology faculty at Oberlin. Each of them spoke about things they really cared about, and I was hooked. I ended up taking lots of biology courses and liked them all. Looking back, three of them had especially lasting impacts. David Egloff taught an ecology course that excited me because there were so many interesting and unanswered questions, and fewer technical details to be mastered than in most science courses. Jim Stewart was a young sabbatical replacement; he taught the evolution course the year I took it. We had two texts, Ernst Mayr’s Populations, Species, and Evolution and Loren Eiseley’s Darwin’s Century. Eiseley’s book traced the antecedents to Darwin’s work as well as the challenges that he faced in his day, and it remains one of my all-time favorites. Last but not least, Dick Levin taught a course on molecular genetics that, looking back, was probably the most important one I took. We used a wonderful text by Gunther Stent that traced the history of that field, focusing on what the questions were as that field got started and how incredibly elegant experiments were devised to answer those questions. The fact that bacteria and viruses were used in so many of those experiments was something I thought about when I was in grad school and pondering my own future directions.DSW: Thanks very much! I love to hear stories about how people became scientists. It is a way of demystifying science. Another fact about you is that you didn’t decide to study microbes until after you had completed your PhD research, which was on insect ecology. This means that in some respects you had to master a completely different set of tools—like a carpenter retraining to become a plumber. Yet, in other respects you were employing the same conceptual tools to study microbes that you did to study insects. Why did you decide to switch and how did your evolutionary training prepare you to make the switch?RL: I never really mastered being a field ecologist. I was good with the concepts and models, but struggled to find a system that was both tractable and interesting to me. Mark-recapture is this wonderful technique conceptually, but getting it to work with beetles in the wild is a lot more difficult. Moreover, my advisor at the University of North Carolina, the late Nelson Hairston, Sr., pushed ecologists to do manipulative experiments, not just uncontrolled observations. That made ecology even more difficult, though it left a lasting impression on how I do science. But I realized that insects, field experiments, and my own skills were not a good match.Also while I was in grad school, I took a course from Janis Antonovics (then at Duke University, now at the University of Virginia) that he called Ecological Genetics. One of his core messages, as he wrote in a thought-provoking paper (1976, Syst. Bot. 1, 233-245), was that “The distinction between ‘ecological time’ and ‘evolutionary time’ is artificial and misleading. Changes of both kinds may be on any time scale: frequently genetic and ecological changes are simultaneous.” That got me really excited with the idea that evolution was not just a museum science about history but an on-going process that could be seen and studied experimentally. Phil Service (a fellow grad student) and I spent long hours discussing how we might integrate ecology and evolution into our future research, and what systems we might use. Phil switched to studying fruit flies for his postdoc, and I thought about that, too. But I remembered the elegant experiments with bacteria and viruses that formed the foundation of molecular genetics, and I thought they should be valuable for evolutionary research, too. So I started reading papers and looking for potential postdoctoral sponsors doing related work. There weren’t many in those days, but one was Bruce Levin (then at the University of Massachusetts, Amherst, now at Emory University). I was particularly interested in work he did with his student Lin Chao (now at the University of California, San Diego) on coevolution between bacteria and the viruses that infect them. I wrote to Bruce about the possibility of working with him, and—lucky for me—he invited me for a visit and then gave me a position, even though I had no experience working with microbes. In fact, Bruce had already retooled twice himself; he started out in human genetics, moved to working on fruit flies, and then as a young faculty member switched to microbes.Despite wanting to make the switch from field ecology, I was also worried: Would I miss working outdoors? Would I find the lab work too difficult? Or maybe boring? In fact, I loved it right away. Not that I was good at it. But I liked the immediacy of the feedback—you spread bacteria on a petri dish one day, and then you count the colonies the next day. And I liked the inferential logic that lay behind much of the work. For example, you don’t see the viruses kill the bacteria by peering through a microscope, but instead you count bacterial colonies before and after exposure to the viruses. And Janis Antonovics was absolutely right about the overlap between ecological and evolutionary time: a single mutation in the bacteria could change their population density in the presence of a virus by several

orders of magnitude, and whether the virus persisted or went extinct depended not only on their own evolution but also on whether bacterial resistance engendered a tradeoff, or cost, in terms of reduced competitive ability.By the way, let me be clear that my switch from the field to the lab is not because I think it’s the better way to do science—it was a personal decision based on my own skills, interests, and affinities. In fact, seeing how hard it is to discover things even in the clean confines of the lab and the simple systems I study only makes me all the more impressed with people like Peter and Rosemary Grant, who observe and make sense of evolution under far more complex and difficult conditions.DSW: Right! Many different types of research are needed. Yet, that doesn’t mean that anything goes. When you say that there weren’t many people like Bruce Levin in those days, and that he had already retooled twice, you are talking about a distinctive approach that the vast majority of microbiologists at that time were not following. You wanted to work with Bruce and not those other people.I often rely upon a famous article by Niko Tinbergen to describe the major conceptual tools of the evolutionary toolkit. As you know, Tinbergen was a pioneer in the study of animal behavior (called Ethology) and shared the Nobel Prize in medicine with Konrad Lorenz and Carl Von Frisch in 1973. Back then, it wasn’t obvious that a behavior such as aggression could evolve by natural selection in the same way as a physical trait such as a deer’s antlers. In his effort to explain how Ethology is a branch of biology, Tinbergen described how four questions need to be asked about all products of evolution, concerning function, history, mechanism, and development. Asking all four questions in conjunction with each other constitutes a fully rounded evolutionary approach. I see your research as exemplifying this approach in an organism that allows for an extraordinary degree of experimental manipulation. Is that (roughly) how you see it?RL: I certainly appreciate the distinction between proximate and ultimate causes. I’d say that I was really more interested in the latter when I started out. But after working so long with a model organism like E. coli, and with the benefit of talented collaborators and students who have different skills and interests, I’ve realized the opportunity to integrate things and delve deeper into mechanisms. One thing that I wrestle with, though, is how we use the term “mechanism”—for many biologists, it seems to mean only molecular or chemical explanations. But I would argue that natural selection is a mechanism, too, it just operates at a different level.DSW: Sure--there’s no reason why the term “mechanism” can’t be used to describe natural selection in certain contexts. I think what Tinbergen was calling attention to with the “Function” question is the ability to reason about the properties of organisms on the basis of adaptation and natural selection, without any reference to their genes or physical makeup. We can confidently predict that many desert species will evolve to be sandy colored to avoid being seen by their predators and prey, regardless of whether they are insects, snails, amphibians, reptiles, birds, or mammals. As long as there is heritable phenotypic variation in coloration, the genes and physical makeup of these very different organisms (chitin for insects, scales for reptiles, feathers for birds, fur for mammals) can be ignored in making this prediction. We call this “Adaptationist thinking” or “Natural Selection thinking”. That’s how Darwin and Wallace could make so much progress developing their theory, knowing nothing about genes and little about the physical makeup of organisms, and it remains one of the most powerful tools in the evolutionary toolkit.RL: I agree completely. When I teach students or give talks to people who hear so much nowadays about genes and DNA, I emphasize that Darwin knew nothing about the material basis of heredity. He knew there was heredity, and he knew that it was important for his theory, but he didn’t know how it worked. And he was open about his ignorance—he wrote in The Origin that “Our ignorance of the laws of variation is profound.” Daniel Dennett said it well that “the process of natural selection is substrate-neutral.” That’s why Darwin and Wallace could begin to make sense of evolution without knowing anything about the biochemical basis of heredity.DSW: This is why you can have a separate line of research on digital organisms, but that’s another story. In the case of your E. coli experiment, you created twelve isolated lines of E. coli and grew them on a liquid culture medium with glucose as the main energy source. You also had the amazing ability to create a frozen fossil record and to bring the ancestors back to life to compare with their own descendants. You could confidently predict that E. coli would evolve an enhanced ability to utilize glucose as an energy source in this environment, as surely as desert animals evolve to be sandy colored. Is that what you found?

RL: That’s right. When I started working with bacteria, one of the obvious appeals was their fast generations. That’s what allows us to see evolution in action. But now I realize that the ability to freeze the bacteria and bring them back to life is an even more important part of our work. It allows us to compare and even directly compete organisms that lived at different times. In effect, we can do time-travel experiments.

And yes, all 12 lines have become much better at using glucose. They grow faster than the ancestor, they’re more efficient, and they outcompete the ancestor in the glucose medium where they evolved. To an evolutionary biologist, these changes aren’t surprising. But it’s a nice, clear, and simple demonstration of adaptation by natural selection in real time.DSW: Now let’s turn to Tinbergen’s “History” question. Even though each of your isolated lines responded to selection, they did so in different ways. After all, genetic mutations are chance events, so it’s extremely unlikely for the exact same mutations to occur in the different lines. How is it possible for each line to evolve the same adaptation in functional terms (ability to utilize glucose) but with different genetic mutations and metabolic pathways?RL: One of the important points you raise is that the changes we see in the LTEE—that’s our shorthand for the long-term evolution experiment—depend entirely on new mutations that have occurred during the experiment. Of course, genetic variation is required for adaptation by natural selection. In most systems, whether fruit flies in the lab or finches in the Galapagos, there’s a lot of genetic variation present at the start of a study. But in the LTEE, all of the changes that we see are fueled by new mutations that arise during the course of the experiment.There certainly are a lot of different mutations that could occur. Just considering point mutations, there are about 14 million possibilities: there are 4.6 million base pairs in the genome of the E. coli strain we use, and there are 3 alternatives in the genetic code at each of those positions. Plus there are deletions, insertions, and other kinds of mutations as well. In fact, though, it’s surprisingly likely that the exact same ones—at least the same point mutations—would occur in all of the populations because the populations are so large. Without going through the math, I’ve estimated that something like a billion point mutations have occurred in each of the 12 populations over the course of the LTEE. That’s a lot more than there are different point mutations, meaning that every possibility has been tried many times over. Having said that, though, most mutations—even most beneficial mutations—are lost due to random genetic drift before they can gain a foothold in an evolving population. Moreover, the mutations don’t occur in the same order in the different populations because of these random effects. And as some beneficial mutations do gain a foothold, rise in abundance, and eventually get fixed in a population, they change the genetic background, or context, in which future mutations arise. So, while the set of one-step mutations may be reasonably finite, at least given the huge size of bacterial populations, the number of possible paths is essentially infinite. I find that really exciting!Getting back to our results about how the bacteria have adapted to glucose, one of the things that’s rewarding about such a long experiment is that new technologies have come along that I never could have imagined. No genome had been sequenced when I started the LTEE, but now we’ve sequenced hundreds of genomes to find the mutations that have accumulated along the way. What we see is a fascinating mix of predictability and unpredictability. On the one hand, we almost never see that the exact same mutation has been fixed in even two of the replicate populations. On the other hand, we see many cases—far more than you’d expect by chance—where the same genes have different mutations in several or even all 12 of the populations. These are cases, then, of parallel evolution, where independently evolving lineages find similar solutions to some environmental challenge.Now you might wonder, maybe these parallel changes are not because selection has favored the phenotypic changes caused by the mutations, but instead those genes are somehow mutational hotspots. That’s plausible, but another nice thing about working with E. coli is that we can dig deeper and test our hypothesis that these parallel mutations confer a benefit. We can make isogenic constructs where we move individual mutations around, keeping everything else the same, and then compete strains with and without a mutation of interest. We’ve done that for some of the genes where we’ve seen parallel changes, and in most cases we’ve confirmed that those mutations provide a measurable advantage under the conditions where they evolved.But against this backdrop of parallelism, or repeatability, the longer the LTEE has been running, the more we see that each population really is following its own path. Some of the differences are subtle, but others are more dramatic. Let me give a few examples:
- The ancestral strain started out with a low mutation rate, but six populations evolved hypermutability, which increases the point-mutation rate by roughly 100-fold.
- One population evolved a stable polymorphism in which two ecologically distinct lineages have coexisted for over 50,000 generations.
- Another population evolved the ability to use a second carbon source, called citrate, which neither the ancestor nor any of the other lineages can use; the citrate is in the medium as a chelating agent, and coli can’t normally grow on it.
- More subtly, one population has been stuck on the lowest rung, so to speak, for a long time; that is, for tens of thousands of generations it has consistently been less fit than any of the other populations.
So what we see is this deep tension between chance and necessity—between the randomness of mutations and genetic drift, on the one hand, and the predictability of natural selection as it discovers similar solutions when replicate populations are confronted with the same environmental challenges. And both sets of forces—the random and the predictable, as it were—together give rise to what we call history.DSW: Amazing! That’s what makes the LTEE so great. You’ve already pointed out that all of your lines started out with a single genetically identical clone. After 2000 generations, you started a second experiment while keeping the first one going. In the second experiment, you switched the main energy source to maltose rather than glucose. What happened and how was adaptation to the new resource different than the first experiment, based on the 2000-generation history of each of the lines?RL: Yes, they all started from the same ancestral strain, except for a genetic marker that was embedded in the experiment to tell lines apart. As part of his dissertation, Mike Travisano (now professor at the University of Minnesota) showed that while the 12 populations were similarly fit when they competed in the same environment as the LTEE, they were much more heterogeneous if they competed in environments with different sugars from the glucose on which they had evolved for, at that time, 2000 generations. One of the alternative sugars was maltose—interestingly enough, that’s di-glucose, but it made a big difference. Some of the populations that evolved in glucose were much less fit than others when they competed for maltose and, in fact, some of them were even less fit than their ancestor. So Mike then asked what would happen if the glucose-evolved lines were given the opportunity to evolve for 1000 generations in the same environment except with maltose instead of glucose as the limiting resource. Would history be destiny? Would the chance differences that arose while they evolved in glucose constrain their future potential in maltose? Or would history be erased, with the laggards catching up to the leaders because they had more or steeper paths to improvement? When Mike ran that experiment, selection dominated history—that is, the laggards almost caught the leaders. So despite the big differences in growth on maltose that arose during the preceding 2000 generations on glucose, after 1000 generations in maltose all the lines had similarly high fitness in that new environment.At the end of that paper, though, we acknowledged that the constraints imposed by history might become more important, or even insurmountable, over a longer time scale. We wrote that “the footprint of history might eventually become too deep to be obscured even by intense selection.” The longer the LTEE runs, the more we see that each population has its own evolutionary path and personality—sometimes the differences are big ones, and sometimes they’re subtle. But it raises the possibility that some populations might eventually get so poor at using maltose that, if moved into a maltose environment, they would go extinct or at least take much longer than 1000 generations to become well adapted to that sugar. In fact, a current student, Jay Bundy, is starting to address exactly that question: how long does it take for the footprint of history to become so deep that it presents a serious barrier to future evolution?DSW: Awesome! Now let’s turn to Tinbergen’s “Mechanism” question. By this he meant that every trait has a physical basis, which must be understood in addition to its functional basis. Returning to my desert coloration example, even if we can ignore the genes and physical makeup of the different species while reasoning in “Adaptation thinking” mode, cryptic coloration has a genetic and physical basis in each and every species. One of the great advantages of working with E. coli is that so much is mechanistically known about it. How have you and your colleagues employed this mechanistic level of understanding in your research?RL: In some cases, we know quite a bit about the mechanisms of how the bacteria have changed and adapted to their environment. For example, we know that the population that gained the new ability to eat citrate required a complex mutation that brought together two segments of DNA, one that encoded a protein and the other that caused the protein to be expressed in a new context. As you say, working with an organism like E. coli—one that’s been studied by hundreds of scientists for decades—provides a reservoir of mechanistic knowledge. Even so, we sometimes find mutations and then, by genetic manipulation, show that the mutated gene is important, but we still don’t understand why functionally that mutation is beneficial. One challenge is that many of the genes that show parallel evolution—the ones we know or strongly suspect are important—have complex, high-level functions. Some are called “global regulators” because they influence many lower-level processes. At first, it was surprising to me that these critical functions changed in an experiment like the LTEE. I suspect what’s going on is these global regulators are integrating and balancing competing demands in the cell, and in the LTEE we’ve removed some demands—there’s no need to resist bile salts or viruses, for example—while making others even more important than in nature—there’s only the glucose to eat, so a cell better get it fast, before the other cells have eaten it all up.
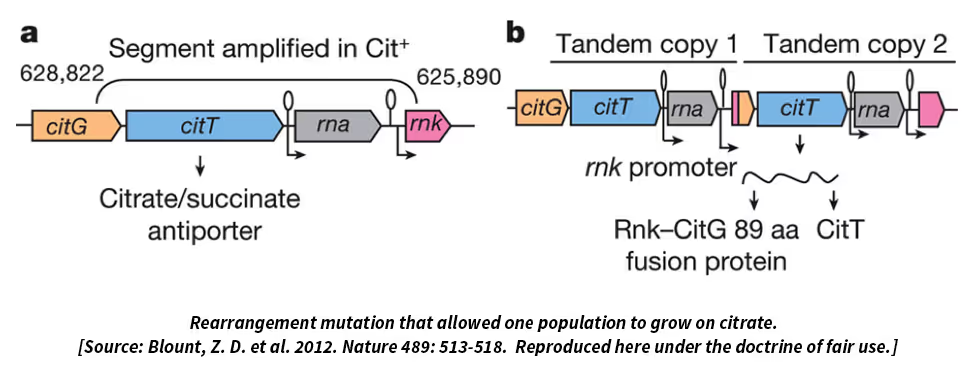
My hope is that we will attract hard-nosed biochemists, physiologists, and molecular biologists to study the LTEE lines, especially as we publish genomic analyses that point to the involvement of particular biochemical and metabolic pathways. I think there’s a lot more still to be learned, even in a well-studied organism like E. coli, by combining reductionist and integrative approaches. It’s important to understand that fitness—reproductive success, if you prefer—is the ultimate arbiter of success in an evolutionary context and the most integrative of all phenotypic traits. In terms of cellular processes, anything and everything could matter, and the processes are all connected, at least indirectly via the biochemical pools and fluxes in cells. So a deeper understanding will require an evolutionary-based systems-level approach, not the “break it and see what happens” approach that’s long been used to atomize organisms into their parts-list.I mentioned Peter and Rosemary Grant’s beautiful work on Darwin’s finches in the Galapagos, and I like to contrast the challenges of their work and ours on the LTEE. Peter and Rosemary are gifted naturalists and can see what matters to the birds in terms of getting food, finding mates, and so on. Through careful observation, they see that traits like the size and shape of the beaks of the finches matter for opening seeds. But much more difficult for them is to show, as they have done, two things: first, that subtle differences in beaks really do affect the survival and reproductive success of the birds; and second, that those differences in beak morphology are, at least in part, inherited. Those are very challenging things to do with birds while working on a remote island, yet they’ve managed to do them. With the E. coli and the LTEE, we have the inverse problem. On the one hand, it’s pretty straightforward to follow lots of bacterial generations and show that the descendants are more fit and replicate faster than their ancestors. And with genome sequencing and genetic manipulations, we can find genetic differences and even identify which mutations matter. But really opening up the “black box”—understanding how functionally a mutation makes one genotype more fit than another—remains a tough problem for us in most cases.DSW: The comparison with the Grants’ research is indeed apt. I know that, through collaborations, they are getting at the proximate mechanisms also. Finally, we get to Tinbergen’s “Development” question, which requires understanding how a given trait comes into being during the lifetime of an organism, in addition to the mechanistic basis of the fully developed trait. Does the concept of development make sense in a bacterium such as E. coli? Does your research include Tinbergen’s fourth question?RL: I guess that depends on definitions. Since E. coli is a single-celled organism, it doesn’t have development in the usual sense. But cells do have a life cycle of sorts and lifetimes over which their circumstances change. Understanding how bacteria get information from their environments and use that information to turn genes on and off has long been a part of microbial genetics, as exemplified by François Jacob and Jacques Monod’s work on the lac operon, which won a Nobel Prize. That work was also very influential in getting people to think about the role of gene regulation in the development of multicellular organisms.Thinking about life histories in the context of the LTEE, every day the bacteria are diluted into fresh medium, and each day they experience a series of growth phases that correspond to different physiological states. There’s a lag phase when the cells are gearing up to start growing again on the renewed resources. That’s followed by exponential growth, when the cell population is growing at its maximum rate for the conditions. But as the cell numbers increase, they draw down the glucose and, once it’s depleted, they enter what’s called stationary phase. They don’t die appreciably during stationary phase, at least not when it’s fairly brief like in the LTEE. Anyhow, we’ve found that the bacteria not only evolved faster exponential growth, they’ve also substantially reduced the duration of the lag phase prior to growth. So they’ve evolved to respond faster to the transition into fresh medium or perhaps to end up in stationary phase better prepared for that coming transition.Also, all 12 lines make cells that are much larger than those of the ancestral strain. That was another surprise and seems counterintuitive in a couple of respects. First, larger organisms usually grow more slowly and have longer generation times than smaller ones. Think of elephants versus mice, for example. Yet in the LTEE we’ve evolved cells that are larger, grow faster, and have shorter generation times than their ancestor. Second, from a functional perspective, if you had asked me at the start of the LTEE whether the bacterial cells would evolve to become smaller or larger, I’d have predicted smaller because that would give them a higher ratio of surface area to volume, which would seem advantageous in acquiring resources. But the bacteria clearly had a different opinion, so to say, because they’ve become much larger—and in most cases they’re a bit rounder and less rod-shaped, which further reduces the area-to-volume ratio. So what’s going on? Once again, I don’t know, but I think the bigger cells might have an advantage because they are bigger sponges. Although their surface-to-volume ratio is less favorable, each big cell has more total surface area than each small cell. In the LTEE there’s a race every day to get as much of the glucose as possible before it runs out. A larger cell can hold onto the glucose molecules that it gobbles up and pass them on to its daughter cells. So maybe the larger cells, while appearing inelegant from an engineering standpoint, are a maternal investment in the kids’ future. That would be pretty cool.DSW: Hmmm. Parental care in bacteria! So far I have been portraying your research as confirming the basic elements of evolutionary theory, as if all of your results could be predicted beforehand. However, I know that empirical research usually also results in surprises that no one could have predicted before hand. You’ve already mentioned the ability to digest citrate, which might be like us evolving the ability to digest hay. What are some other big surprises?RL: The results are certainly consistent with the basic elements of evolutionary theory. Those elements are pretty darn simple and well understood: mutation and recombination generate heritable variation, and selection and drift affect the fate of variants. (Although the bacteria in the LTEE don’t have recombination in the sense of genetic exchange.) Yet from “this view of life”—to echo Darwin’s words and your site’s name—“endless forms most beautiful and most wonderful have been, and are being, evolved.”It might seem depressing, from the vantage of scientific discovery, to say that all of evolution comes down to a few basic processes. But physics, too, boils down to just a few fundamental forces—gravity, electromagnetism, and strong and weak nuclear interactions. Yet together they gave rise to the elements and chemistry as well as to stars, galaxies, and solar systems. In the same vein, the core processes of organic evolution produce fascinating higher-level dynamics and phenomena like speciation and cooperation.Now back to some surprises from the LTEE that could not have been predicted, or at least weren’t what I expected at the outset. I’m tempted to say that, beyond the fact that the bacteria got more fit, almost everything has been unpredictable—certainly the details of what changed genetically, but also many of the interesting phenotypic changes. I’ve mentioned several of them: the evolution of higher mutation rates, the larger cells, the stable polymorphism despite the simplicity of the environment, and the evolution of the ability to use citrate. On that last point, perhaps the surprising thing has gone from “Wow, it happened!” to “Gee, why is it so difficult that only one population has figured it out even after 60,000 generations?”At a more abstract level, I’ve been surprised by two other outcomes. When I started the LTEE, I thought the replicate populations would follow very different paths and that their divergence would be obvious just from looking at the fitness trajectories. In the metaphor of Sewall Wright, I imagined the adaptive landscape was rugged with multiple peaks, such that the populations would reach different fitness levels based on the luck o

f their early mutational steps. As I said earlier, it is the case that the longer we watch the LTEE, the more we see that the populations have diverged in terms of both their fitness and genetics. Still, I’ve been surprised that the fitness trajectories are so similar and that there’s been so much genetic parallelism.Even more surprising to me, I thought the fitness trajectories would level off—that the bacteria would reach an asymptote, or upper limit, beyond which they couldn’t grow any faster. We’ve seen the rate of improvement in fitness slow over time, but it slows down in a way that fits a power-law relation, which has no upper limit but instead tracks the logarithm of time. Not only does the power-law model fit the data much better than a model with an asymptote, we’ve shown that it is predictive. For example, given just 5000 generations of fitness data, the power-law model predicts the trajectory very well out to 50,000 generations. Now there may well be some ultimate limit, but even if we extrapolate the trajectory to millions of generations, the implied growth rates and fitness effects that would be subject to selection are biologically reasonable and plausible. Evolutionary biologists have usually thought of changing environments, including coevolution with other species, as what keeps evolution going. Of course, changing environments are a huge part of nature and critically important for understanding evolution. But our results also suggest that evolution can keep discovering ways to improve even in a constant world. I hope the LTEE will continue for many more generations—and here I mean generations of scientists—to see whether this prediction holds up and to see what other surprises emerge from the potent mix of bacteria, time, and human ingenuity.DSW: This has been a blast! I think that our conversation has done a good job describing evolutionary theory as a workaday toolkit--capable of producing a cathedral of knowledge.RL: Yes, it’s been great fun! And you’ve made me think about how our work relates to Tinbergen’s framework for understanding evolution.




